# run these every time you open Rstudio
library(tidyverse)
library(oibiostat)
library(janitor)
library(rstatix)
library(knitr)
library(gtsummary)
library(moderndive)
library(gt)
library(broom)
library(here)
library(pwr) # new-ishDay 14: Comparing Means with ANOVA (Section 5.5)
BSTA 511/611
Load packages
- Packages need to be loaded every time you restart R or render an Qmd file
- You can check whether a package has been loaded or not
- by looking at the Packages tab and
- seeing whether it has been checked off or not
Goals for today (Section 5.5)
Analysis of Variance (ANOVA)
When to use an ANOVA
Hypotheses
ANOVA table
Different sources of variation in ANOVA
ANOVA conditions
F-distribution
Post-hoc testing of differences in means
Running an ANOVA in R
Disability Discrimination Example
- The U.S. Rehabilitation Act of 1973 prohibited discrimination against people with physical disabilities.
- The act defined a disabled person as any individual who has a physical or mental impairment that limits the person’s major life activities.
- A 1980’s study examined whether physical disabilities affect people’s perceptions of employment qualifications
- Researchers prepared recorded job interviews, using same actors and script each time.
- Only difference: job applicant appeared with different disabilities.
- No disability
- Leg amputation
- Crutches
- Hearing impairment
- Wheelchair confinement
- 70 undergrad students were randomly assigned to view one of the videotapes,
- then rated the candidate’s qualifications on a 1-10 scale.
- The research question: are qualifications evaluated differently depending on the applicant’s presented disability?
Load interview data from .txt file
.txtfiles are usually tab-deliminated files.csvfiles are comma-separated files
read_delimis from thereadrpackage, just likeread_csv
employ <- read_delim(
file = here::here("data", "DisabilityEmployment.txt"),
delim = "\t", # tab delimited
trim_ws = TRUE)trim_ws: Should leading and trailing whitespace be trimmed from each field before parsing it?
summary(employ) disability score
Length:70 Min. :1.400
Class :character 1st Qu.:3.700
Mode :character Median :5.050
Mean :4.929
3rd Qu.:6.100
Max. :8.500 employ %>% tabyl(disability) disability n percent
amputee 14 0.2
crutches 14 0.2
hearing 14 0.2
none 14 0.2
wheelchair 14 0.2Factor variable: Make disability variable a factor variable
glimpse(employ)Rows: 70
Columns: 2
$ disability <chr> "none", "none", "none", "none", "none", "none", "none", "no…
$ score <dbl> 1.9, 2.5, 3.0, 3.6, 4.1, 4.2, 4.9, 5.1, 5.4, 5.9, 6.1, 6.7,…summary(employ) disability score
Length:70 Min. :1.400
Class :character 1st Qu.:3.700
Mode :character Median :5.050
Mean :4.929
3rd Qu.:6.100
Max. :8.500 Make disability a factor variable:
employ <- employ %>%
mutate(disability = factor(disability))What’s different now?
glimpse(employ)Rows: 70
Columns: 2
$ disability <fct> none, none, none, none, none, none, none, none, none, none,…
$ score <dbl> 1.9, 2.5, 3.0, 3.6, 4.1, 4.2, 4.9, 5.1, 5.4, 5.9, 6.1, 6.7,…summary(employ) disability score
amputee :14 Min. :1.400
crutches :14 1st Qu.:3.700
hearing :14 Median :5.050
none :14 Mean :4.929
wheelchair:14 3rd Qu.:6.100
Max. :8.500 Factor variable: Change order & name of disability levels
What are the current level names and order?
levels(employ$disability)[1] "amputee" "crutches" "hearing" "none" "wheelchair"What changes are being made below?
employ <- employ %>%
mutate(
# make "none" the first level
# by only listing the level none, all other levels will be in original order
disability = fct_relevel(disability, "none"),
# change the level name amputee to amputation
disability = fct_recode(disability, amputation = "amputee")
)fct_relevel()andfct_recode()are from theforcatspackage: https://forcats.tidyverse.org/index.html.forcatsis loaded withlibrary(tidyverse).
New order & names:
levels(employ$disability) # note the new order and new name[1] "none" "amputation" "crutches" "hearing" "wheelchair"Data viz
- What are the
scoredistribution shapes within each group? - Any unusual values?
ggplot(employ, aes(x=score)) +
geom_density() +
facet_wrap(~ disability)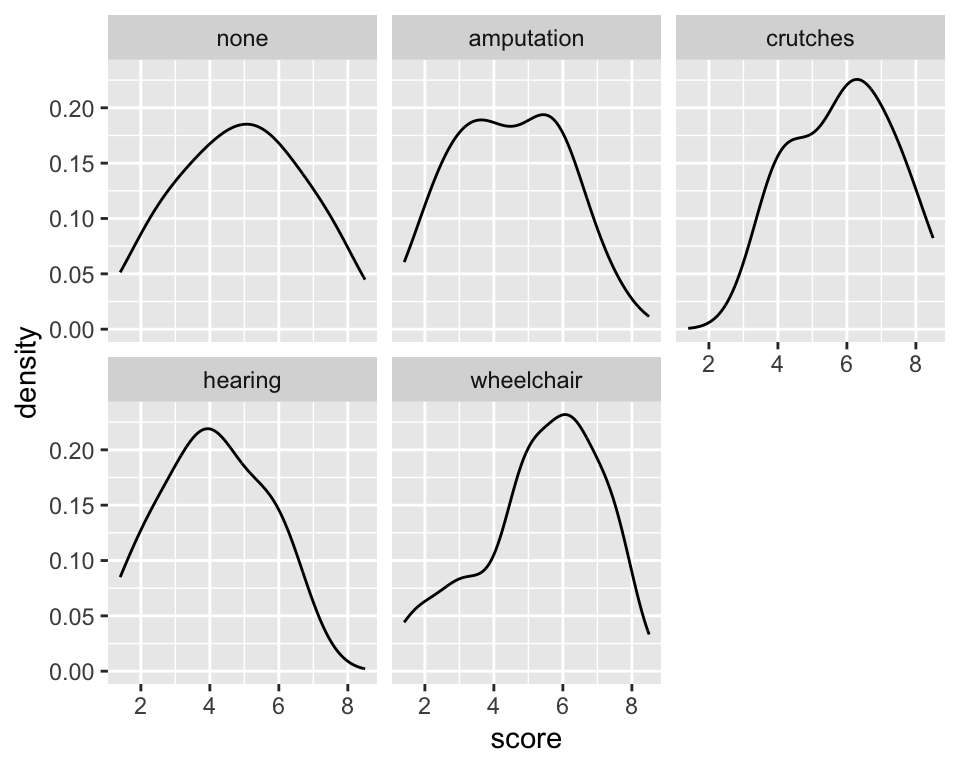
library(ggridges)
ggplot(employ,
aes(x=score,
y = disability,
fill = disability)) +
geom_density_ridges(alpha = 0.4) +
theme(legend.position="none")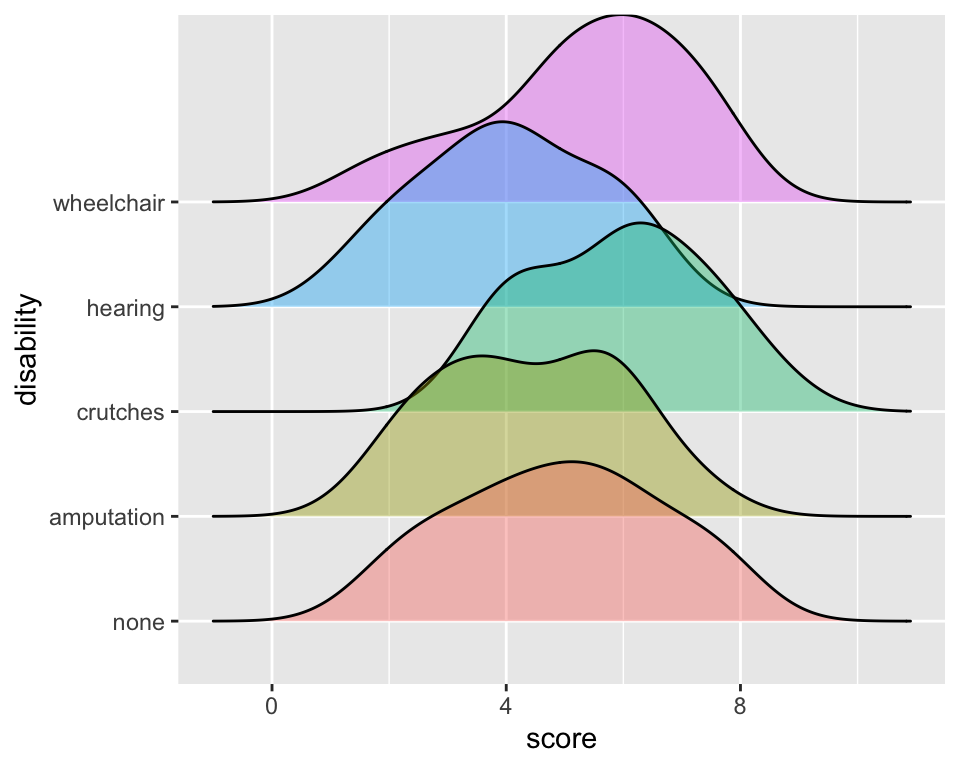
- Compare the
scoremeasures of center and spread between the groups
ggplot(employ, aes(y=score, x = disability,
fill = disability)) +
geom_boxplot(alpha =.3) +
coord_flip() +
geom_jitter(width =.1, alpha = 0.3) +
theme(legend.position = "none")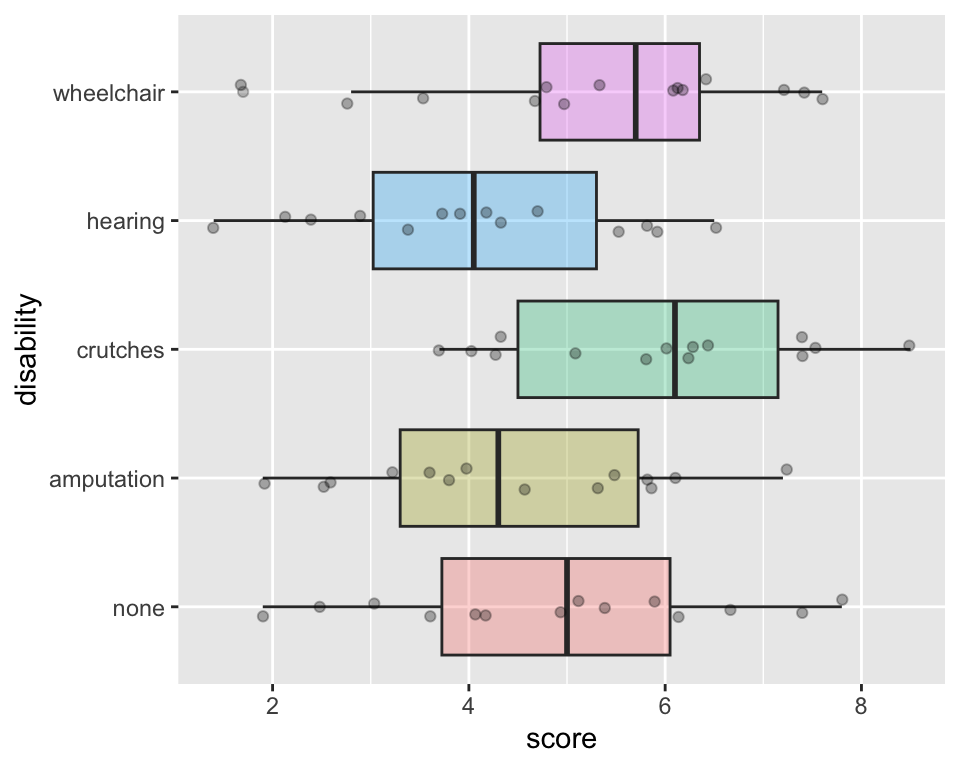
ggplot(employ, aes(x = disability, y=score,
fill=disability, color=disability)) +
geom_dotplot(binaxis = "y", alpha =.5) +
geom_hline(aes(yintercept = mean(score)),
lty = "dashed") +
stat_summary(fun ="mean", geom="point",
size = 3, color = "grey33", alpha =1) +
theme(legend.position = "none")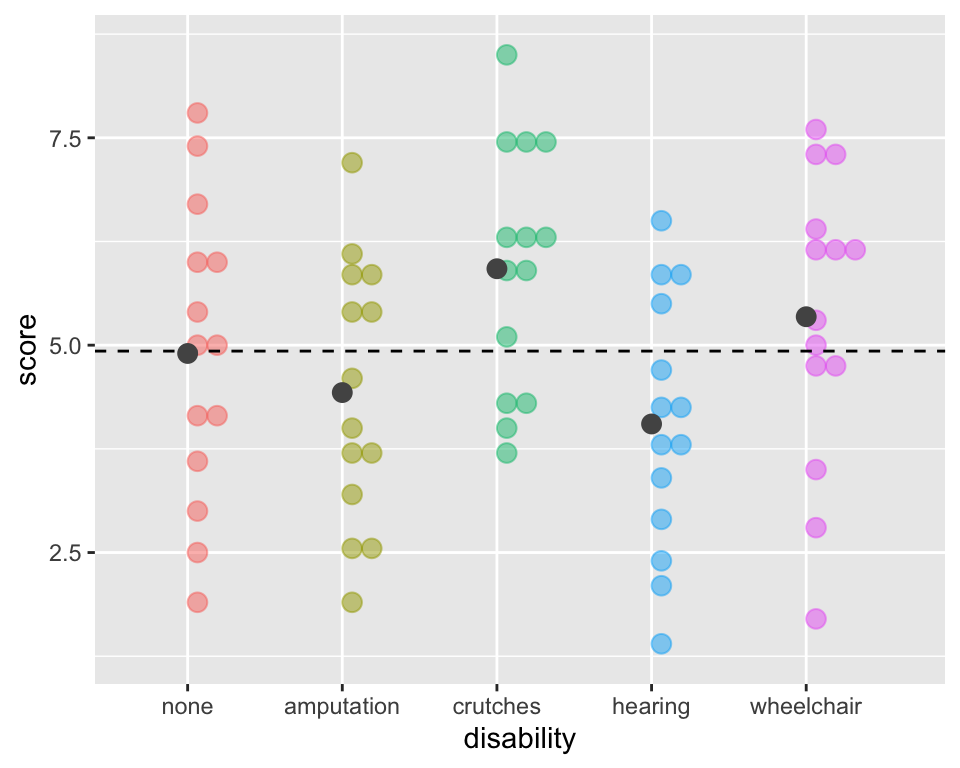
ANOVA
Hypotheses
To test for a difference in means across k groups:
\[\begin{align} H_0 &: \mu_1 = \mu_2 = ... = \mu_k\\ \text{vs. } H_A&: \text{At least one pair } \mu_i \neq \mu_j \text{ for } i \neq j \end{align}\]
Hypothetical examples:
In which set (A or B) do you believe the evidence will be stronger that at least one population differs from the others?
See slides
Comparing means
Whether or not two means are significantly different depends on:
- How far apart the means are
- How much variability there is within each group
Questions:
- How to measure variability between groups?
- How to measure variability within groups?
- How to compare the two measures of variability?
- How to determine significance?
ANOVA in base R
- There are several options to run an ANOVA model in R
- Two most common are
lmandaovlm= linear model; will be using frequently in BSTA 512
lm(score ~ disability, data = employ) %>% anova()Analysis of Variance Table
Response: score
Df Sum Sq Mean Sq F value Pr(>F)
disability 4 30.521 7.6304 2.8616 0.03013 *
Residuals 65 173.321 2.6665
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1aov(score ~ disability, data = employ) %>% summary() Df Sum Sq Mean Sq F value Pr(>F)
disability 4 30.52 7.630 2.862 0.0301 *
Residuals 65 173.32 2.666
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Hypotheses:
\[\begin{align} H_0 &: \mu_{none} = \mu_{amputation} = \mu_{crutches} = \mu_{hearing} = \mu_{wheelchair}\\ \text{vs. } H_A&: \text{At least one pair } \mu_i \neq \mu_j \text{ for } i \neq j \end{align}\]
Do we reject or fail to reject \(H_0\)?
ANOVA tables
See slides
ANOVA: Analysis of Variance
Analysis of Variance (ANOVA) compares the variability between groups to the variability within groups
\[\sum_{i = 1}^k \sum_{j = 1}^{n_i}(x_{ij} -\bar{x})^2 \ \ = \ \sum_{i = 1}^k n_i(\bar{x}_{i}-\bar{x})^2 \ \ + \ \ \sum_{i = 1}^k\sum_{j = 1}^{n_i}(x_{ij}-\bar{x}_{i})^2\]
Notation
- k groups
- \(n_i\) observations in each of the k groups
- Total sample size is \(N=\sum_{i=1}^{k}n_i\)
- \(\bar{x}_{i}\) = mean of observations in group i
- \(\bar{x}\) = mean of all observations
- \(s_{i}\) = sd of observations in group i
- \(s\) = sd of all observations
| Observation | i = 1 | i = 2 | i = 3 | \(\ldots\) | i = k | overall |
|---|---|---|---|---|---|---|
| j = 1 | \(x_{11}\) | \(x_{21}\) | \(x_{31}\) | \(\ldots\) | \(x_{k1}\) | |
| j = 2 | \(x_{12}\) | \(x_{22}\) | \(x_{32}\) | \(\ldots\) | \(x_{k2}\) | |
| j = 3 | \(x_{13}\) | \(x_{23}\) | \(x_{33}\) | \(\ldots\) | \(x_{k3}\) | |
| j = 4 | \(x_{14}\) | \(x_{24}\) | \(x_{34}\) | \(\ldots\) | \(x_{k4}\) | |
| \(\vdots\) | \(\vdots\) | \(\vdots\) | \(\vdots\) | \(\ddots\) | \(\vdots\) | |
| j = \(n_i\) | \(x_{1n_1}\) | \(x_{2n_2}\) | \(x_{3n_3}\) | \(\ldots\) | \(x_{kn_k}\) | |
| Means | \(\bar{x}_{1}\) | \(\bar{x}_{2}\) | \(\bar{x}_{3}\) | \(\ldots\) | \(\bar{x}_{k}\) | \(\bar{x}\) |
| Variance | \({s}^2_{1}\) | \({s}^2_{2}\) | \({s}^2_{3}\) | \(\ldots\) | \({s}^2_{k}\) | \({s}^2\) |
Total Sums of Squares Visually
ggplot(employ, aes(x = disability, y=score,
fill = disability, color = disability)) +
geom_dotplot(binaxis = "y", alpha =.9) +
geom_hline(aes(yintercept = mean(score)),
lty = "dashed") +
# stat_summary(fun = "mean", geom = "point",
# size = 3, color = "grey33", alpha =1) +
theme(legend.position = "none")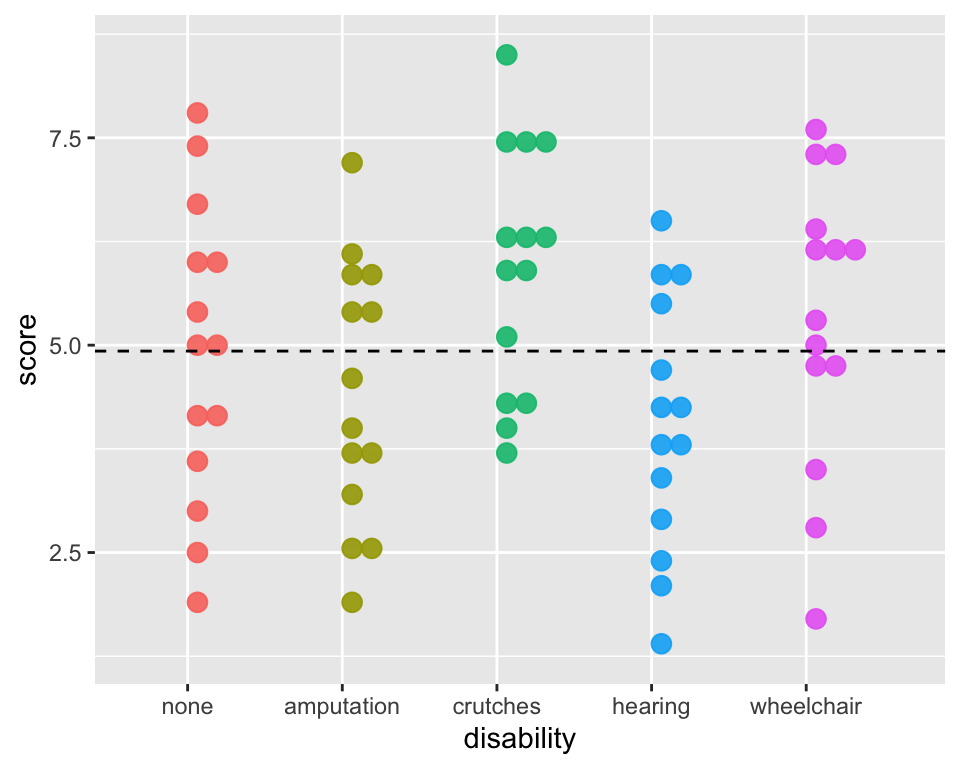
Total Sums of Squares:
\[SST = \sum_{i = 1}^k \sum_{j = 1}^{n_i}(x_{ij} -\bar{x})^2 = (N-1)s^2\]
where
- \(N=\sum_{i=1}^{k}n_i\) is the total sample size and
- \(s^2\) is the grand standard deviation of all the observations
This is the sum of the squared differences between each observed \(x_{ij}\) value and the grand mean, \(\bar{x}\).
That is, it is the total deviation of the \(x_{ij}\)’s from the grand mean.
Calculate Total Sums of Squares
Total Sums of Squares:
\[SST = \sum_{i = 1}^k \sum_{j = 1}^{n_i}(x_{ij} -\bar{x})^2 = (N-1)s^2\]
- where
- \(N=\sum_{i=1}^{k}n_i\) is the total sample size and
- \(s^2\) is the grand standard deviation of all the observations
Total sample size \(N\):
(Ns <- employ %>% group_by(disability) %>% count())# A tibble: 5 × 2
# Groups: disability [5]
disability n
<fct> <int>
1 none 14
2 amputation 14
3 crutches 14
4 hearing 14
5 wheelchair 14\(SST\):
(SST <- (sum(Ns$n) - 1) * sd(employ$score)^2)[1] 203.8429Sums of Squares due to Groups Visually (“between” groups)
ggplot(employ, aes(x = disability, y=score,
fill = disability, color = disability)) +
geom_dotplot(binaxis = "y", alpha =.2) +
geom_hline(aes(yintercept = mean(score)),
lty = "dashed") +
stat_summary(fun = "mean", geom = "point",
size = 3, color = "grey33", alpha =1) +
theme(legend.position = "none")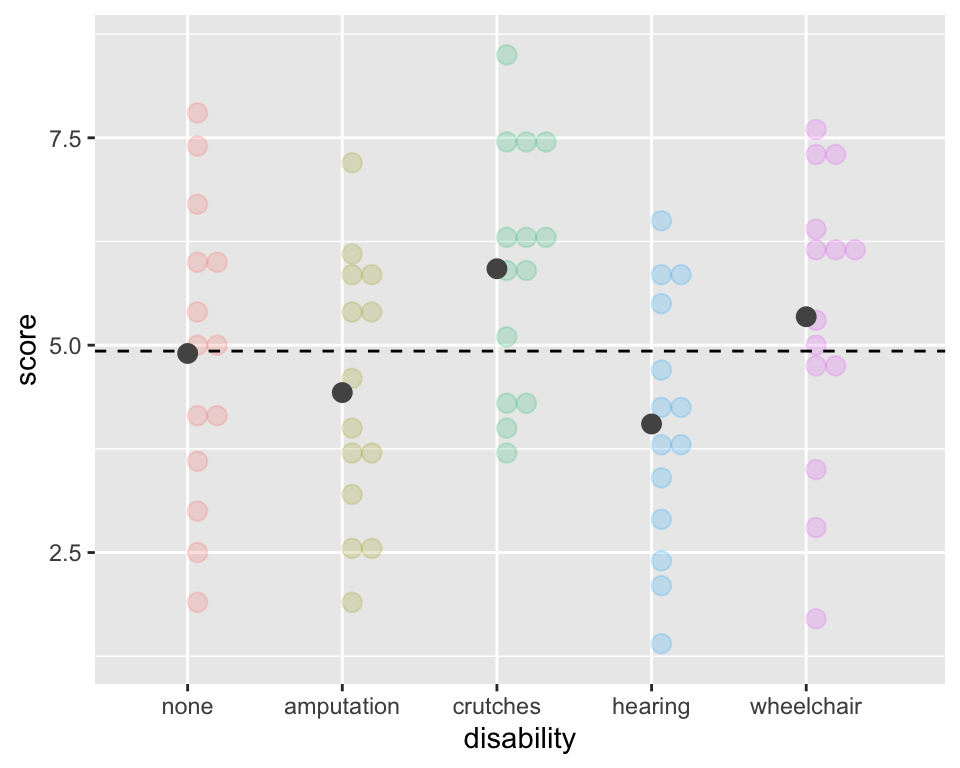
Sums of Squares due to Groups:
\[SSG = \sum_{i = 1}^k n_i(\bar{x}_{i}-\bar{x})^2\]
This is the sum of the squared differences between each group mean, \(\bar{x}_{i}\), and the grand mean, \(\bar{x}\).
That is, it is the deviation of the group means from the grand mean.
Also called the Model SS, or \(SS_{model}.\)
Calculate Sums of Squares due to Groups (“between” groups)
\[SSG = \sum_{i = 1}^k n_i(\bar{x}_{i}-\bar{x})^2\]
ggplot(employ, aes(x = disability, y=score,
fill = disability, color = disability)) +
geom_dotplot(binaxis = "y", alpha =.2) +
geom_hline(aes(yintercept = mean(score)),
lty = "dashed") +
stat_summary(fun = "mean", geom = "point",
size = 3, color = "grey33", alpha =1) +
theme(legend.position = "none")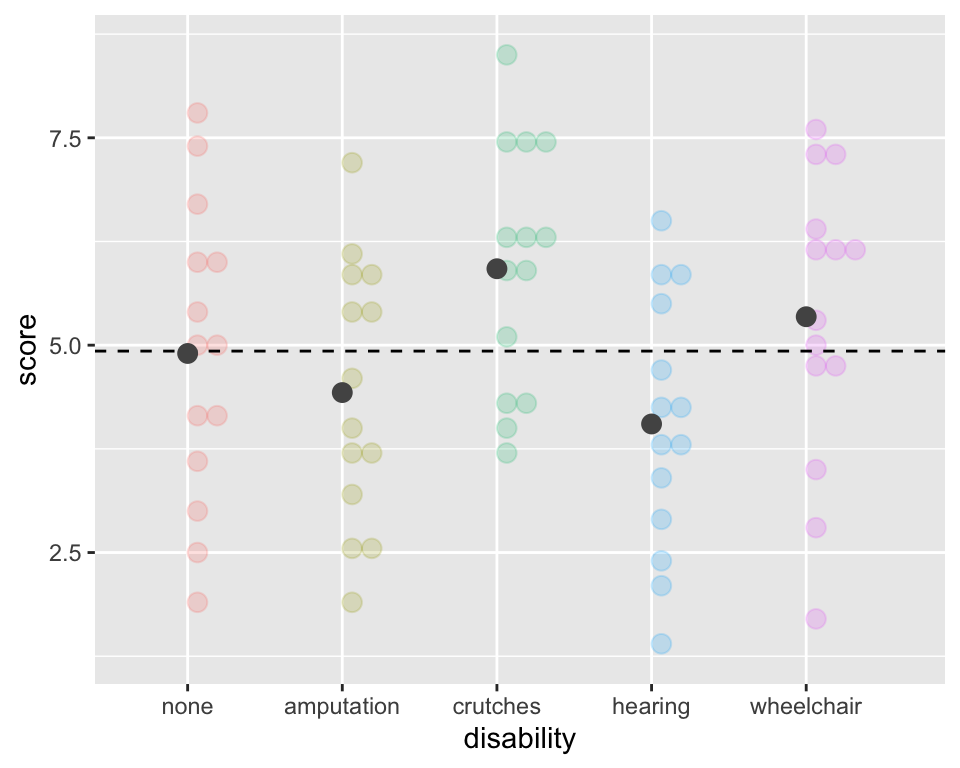
Calculate means \(\bar{x}_i\) for each group:
xbar_groups <- employ %>%
group_by(disability) %>%
summarise(mean = mean(score))
xbar_groups# A tibble: 5 × 2
disability mean
<fct> <dbl>
1 none 4.9
2 amputation 4.43
3 crutches 5.92
4 hearing 4.05
5 wheelchair 5.34Calculate \(SSG\):
(SSG <- sum(Ns$n *
(xbar_groups$mean - mean(employ$score))^2))[1] 30.52143Sums of Squares Error Visually (within groups)
ggplot(employ, aes(x = disability, y=score,
fill = disability, color = disability)) +
geom_dotplot(binaxis = "y", alpha =.5) +
# geom_hline(aes(yintercept = mean(score)),
# lty = "dashed") +
stat_summary(fun = "mean", geom = "point",
size = 3, color = "grey33", alpha =1) +
theme(legend.position = "none")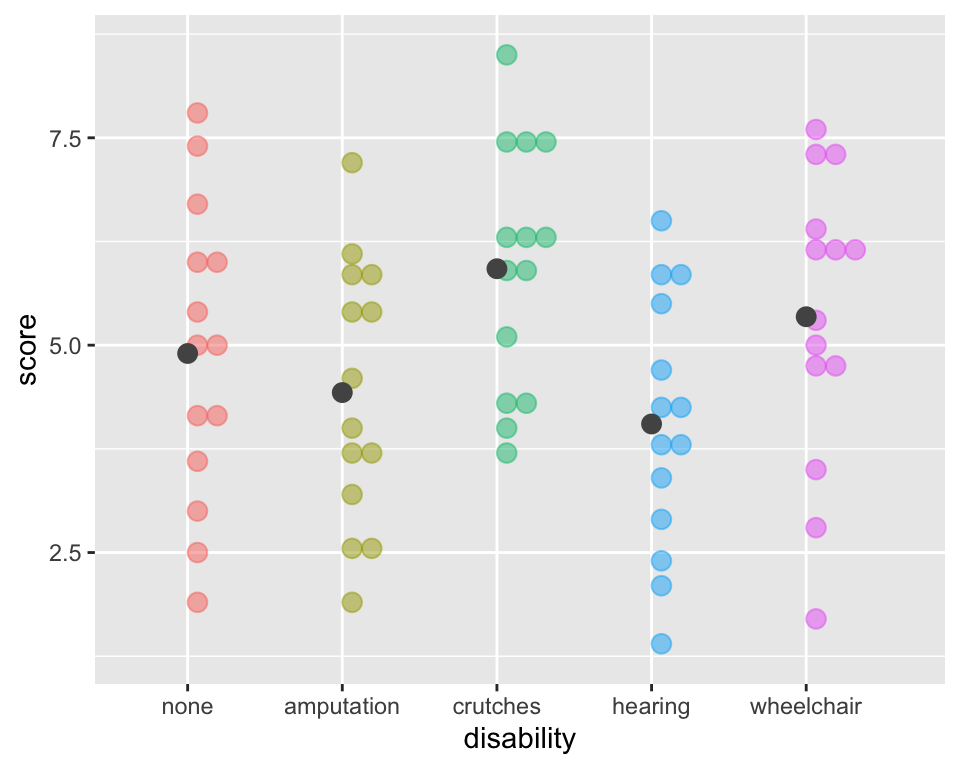
Sums of Squares Error:
\[SSE = \sum_{i = 1}^k\sum_{j = 1}^{n_i}(x_{ij}-\bar{x}_{i})^2 = \sum_{i = 1}^k(n_i-1)s_{i}^2\] where \(s_{i}\) is the standard deviation of the \(i^{th}\) group
This is the sum of the squared differences between each observed \(x_{ij}\) value and its group mean \(\bar{x}_{i}\).
That is, it is the deviation of the \(x_{ij}\)’s from the predicted score by group.
Also called the residual sums of squares, or \(SS_{residual}.\)
Calculate Sums of Squares Error (within groups)
\[SSE = \sum_{i = 1}^k\sum_{j = 1}^{n_i}(x_{ij}-\bar{x}_{i})^2 = \sum_{i = 1}^k(n_i-1)s_{i}^2\] where \(s_{i}\) is the standard deviation of the \(i^{th}\) group
ggplot(employ, aes(x = disability, y=score,
fill = disability, color = disability)) +
geom_dotplot(binaxis = "y", alpha =.5) +
# geom_hline(aes(yintercept = mean(score)),
# lty = "dashed") +
stat_summary(fun = "mean", geom = "point",
size = 3, color = "grey33", alpha =1) +
theme(legend.position = "none")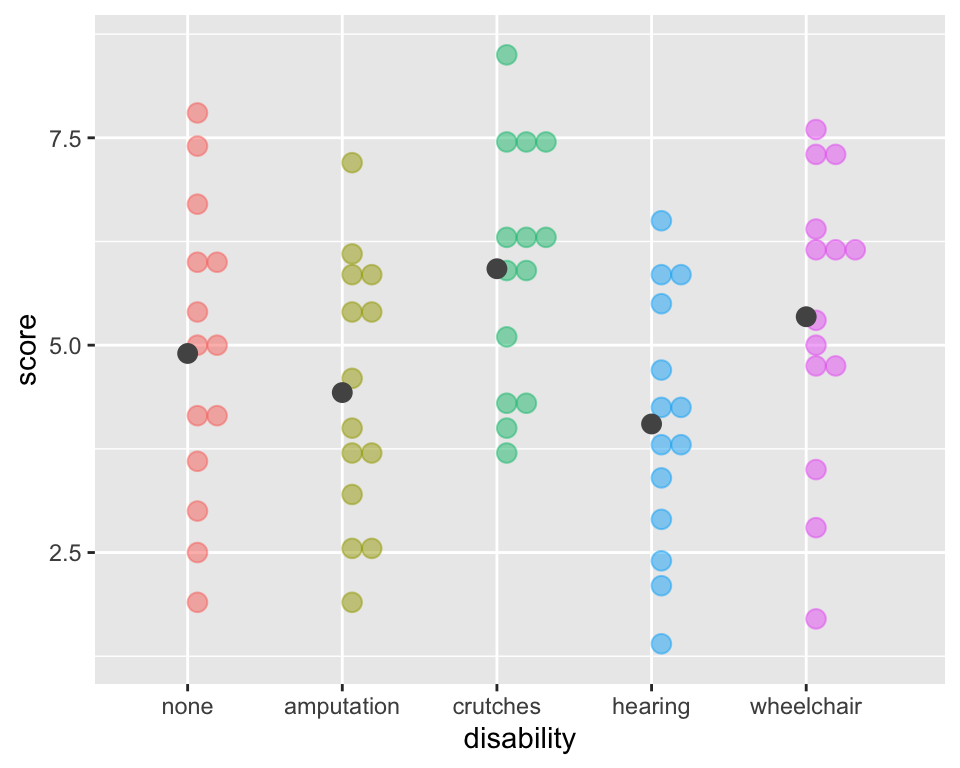
Calculate sd’s \(s_i\) for each group:
sd_groups <- employ %>%
group_by(disability) %>%
summarise(SD = sd(score))
sd_groups# A tibble: 5 × 2
disability SD
<fct> <dbl>
1 none 1.79
2 amputation 1.59
3 crutches 1.48
4 hearing 1.53
5 wheelchair 1.75Calculate \(SSE\):
(SSE <- sum(
(Ns$n-1)*sd_groups$SD^2))[1] 173.3214Verify SST = SSG + SSE
ANOVA compares the variability between groups to the variability within groups
\[\sum_{i = 1}^k \sum_{j = 1}^{n_i}(x_{ij} -\bar{x})^2 \ \ = \ \ n_i\sum_{i = 1}^k(\bar{x}_{i}-\bar{x})^2 \ \ + \ \ \sum_{i = 1}^k\sum_{j = 1}^{n_i}(x_{ij}-\bar{x}_{i})^2\]
\[(N-1)s^2 \ \ = \ \sum_{i = 1}^k n_i(\bar{x}_{i}-\bar{x})^2 \ \ + \ \ \sum_{i = 1}^k(n_i-1)s_{i}^2\]
SST[1] 203.8429SSG + SSE[1] 203.8429Thinking about the F-statistic
If the groups are actually different, then which of these is more accurate?
- The variability between groups should be higher than the variability within groups
- The variability within groups should be higher than the variability between groups
If there really is a difference between the groups, we would expect the F-statistic to be which of these:
- Higher than we would observe by random chance
- Lower than we would observe by random chance
\[F = \frac{MSG}{MSE}\]
ANOVA in base R
# Note that I'm saving the tidy anova table
# Will be pulling p-value from this on future slide
empl_lm <- lm(score ~ disability, data = employ) %>%
anova() %>%
tidy()
empl_lm %>% gt()| term | df | sumsq | meansq | statistic | p.value |
|---|---|---|---|---|---|
| disability | 4 | 30.52143 | 7.630357 | 2.86158 | 0.03012686 |
| Residuals | 65 | 173.32143 | 2.666484 | NA | NA |
Hypotheses:
\[\begin{align} H_0 &: \mu_{none} = \mu_{amputation} = \mu_{crutches} = \mu_{hearing} = \mu_{wheelchair}\\ \text{vs. } H_A&: \text{At least one pair } \mu_i \neq \mu_j \text{ for } i \neq j \end{align}\]
Do we reject or fail to reject \(H_0\)?
Conclusion to hypothesis test
\[\begin{align} H_0 &: \mu_{none} = \mu_{amputation} = \mu_{crutches} = \mu_{hearing} = \mu_{wheelchair}\\ \text{vs. } H_A&: \text{At least one pair } \mu_i \neq \mu_j \end{align}\]
empl_lm # tidy anova output# A tibble: 2 × 6
term df sumsq meansq statistic p.value
<chr> <int> <dbl> <dbl> <dbl> <dbl>
1 disability 4 30.5 7.63 2.86 0.0301
2 Residuals 65 173. 2.67 NA NA Pull the p-value using base R:
round(empl_lm$p.value[1],2)[1] 0.03Pull the p-value using tidyverse:
empl_lm %>%
filter(term == "disability") %>%
pull(p.value) %>%
round(2)[1] 0.03- Use \(\alpha\) = 0.05.
- Do we reject or fail to reject \(H_0\)?
Conclusion statement:
- There is sufficient evidence that at least one of the disability groups has a mean employment score statistically different from the other groups. ( \(p\)-value = 0.03).
Conditions for ANOVA
IF ALL of the following conditions hold:
- The null hypothesis is true
- Sample sizes in each group group are large (each \(n \ge 30\))
- OR the data are relatively normally distributed in each group
- Variability is “similar” in all group groups:
- Is the within group variability about the same for each group?
- As a rough rule of thumb, this condition is violated if the standard deviation of one group is more than double the standard deviation of another group
THEN the sampling distribution of the
F-statistic is an F-distribution
Checking the equal variance condition:
sd_groups # previously defined# A tibble: 5 × 2
disability SD
<fct> <dbl>
1 none 1.79
2 amputation 1.59
3 crutches 1.48
4 hearing 1.53
5 wheelchair 1.75max(sd_groups$SD) / min(sd_groups$SD)[1] 1.210425Testing variances (Condition 3)
Bartlett’s test for equal variances
- \(H_0:\) population variances of group levels are equal
- \(H_A:\) population variances of group levels are NOT equal
Note: \(H_A\) is same as saying that at least one of the group levels has a different variance
- Bartlett’s test assumes the data in each group are normally distributed.
- Do not use if data do not satisfy the normality condition.
bartlett.test(score ~ disability, data = employ)
Bartlett test of homogeneity of variances
data: score by disability
Bartlett's K-squared = 0.7016, df = 4, p-value = 0.9511Levene’s test for equality of variances is not as restrictive: see https://www.statology.org/levenes-test-r/
The F-distribution
- The F-distribution is skewed right.
- The F-distribution has two different degrees of freedom:
- one for the numerator of the ratio (k – 1) and
- one for the denominator (N – k)
- \(p\)-value
- is always the upper tail
- (the area as extreme or more extreme)
F_stat <- 2.86158
ggplot(data.frame(x = c(0, 6)), aes(x = x)) + # set x-axis bounds from 0 to 6
geom_vline(xintercept = F_stat, color = "red") +
# fun = df is specifying (d)ensity of (f) distribution
stat_function(fun = df, args = list(df1=4, df2=65), color = "cornflowerblue") +
geom_area(stat = "function", fun = df, args = list(df1=4, df2=65), fill = "red", alpha =0.3, xlim = c(F_stat, 5)) +
annotate("text", x = 3.5, y = .1, label = "p-value", size=6) 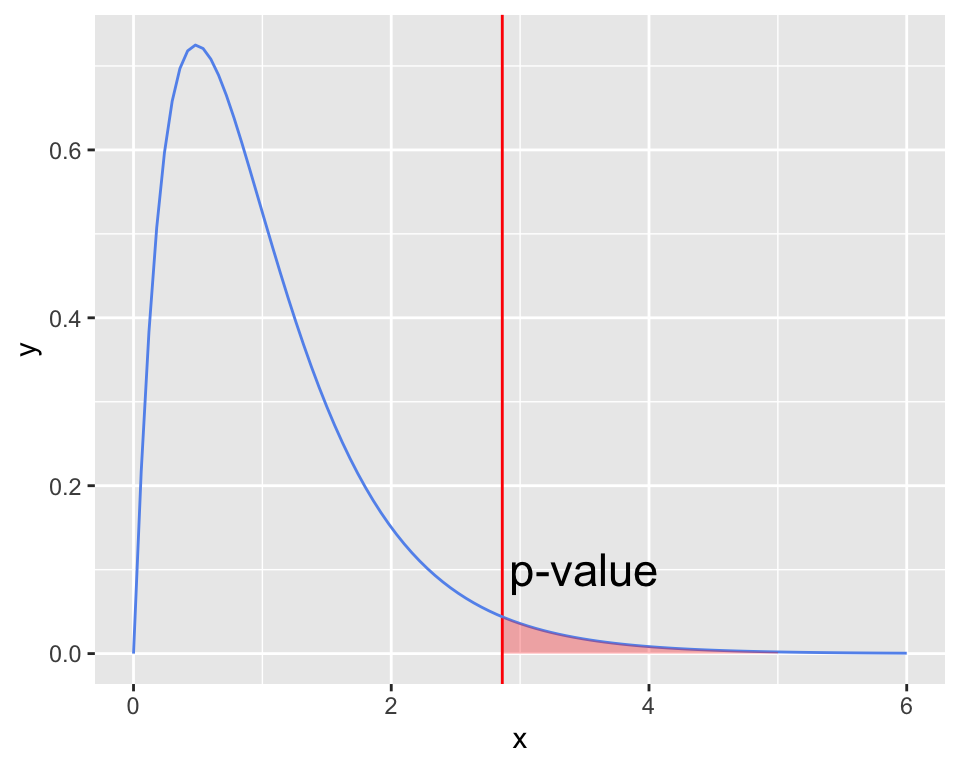
empl_lm %>% gt()| term | df | sumsq | meansq | statistic | p.value |
|---|---|---|---|---|---|
| disability | 4 | 30.52143 | 7.630357 | 2.86158 | 0.03012686 |
| Residuals | 65 | 173.32143 | 2.666484 | NA | NA |
# p-value using F-distribution
pf(2.86158, df1=5-1, df2=70-5,
lower.tail = FALSE)[1] 0.03012688Which groups are statistically different?
- So far we’ve only determined that at least one of the groups is different from the others,
- but we don’t know which.
- What’s your guess?
ggplot(employ, aes(x = disability, y=score,
fill = disability, color = disability)) +
geom_dotplot(binaxis = "y", alpha =.5) +
geom_hline(aes(yintercept = mean(score)),
lty = "dashed") +
stat_summary(fun = "mean", geom = "point",
size = 3, color = "grey33", alpha =1) +
theme(legend.position = "none")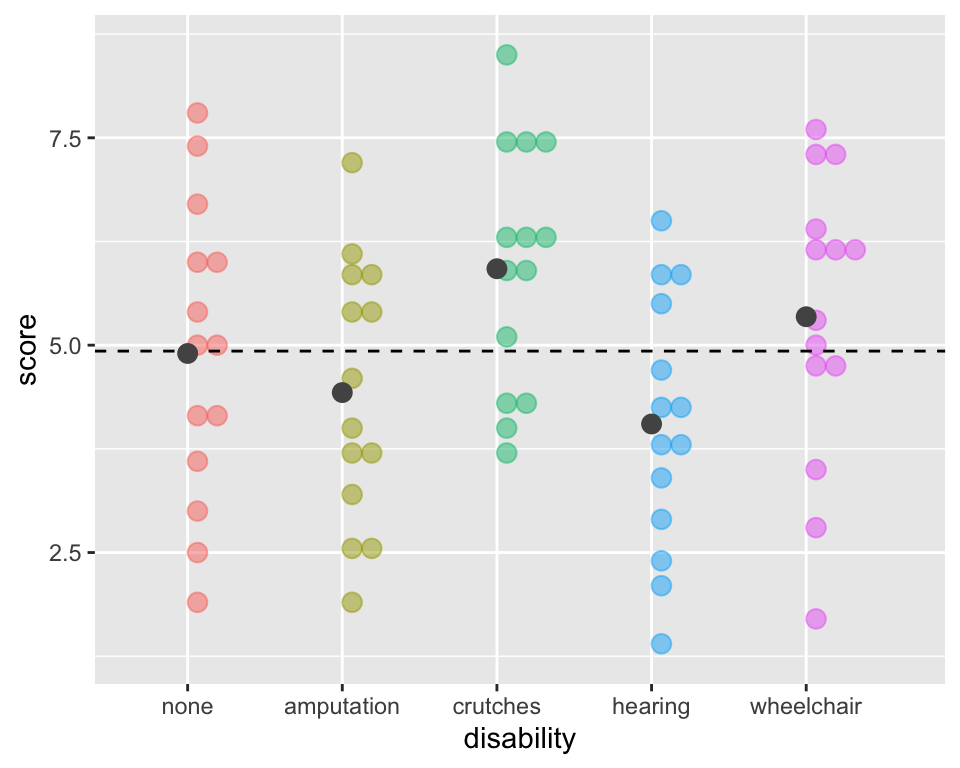
Post-hoc testing for ANOVA
determining which groups are statistically different
Post-hoc testing: pairwise t-tests
- In post-hoc testing we run all the pairwise t-tests comparing the means from each pair of groups.
- With 5 groups, this involves doing \({5 \choose 2} = \frac{5!}{2!3!} = \frac{5\cdot 4}{2}= 10\) different pairwise tests.
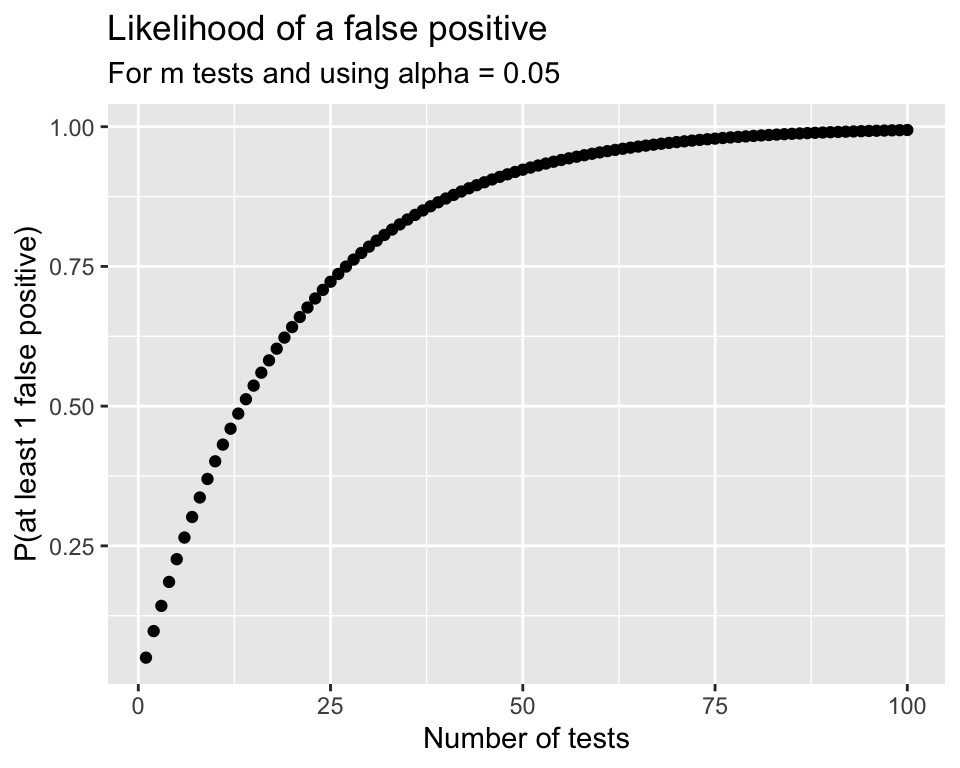
Problem:
- Although the ANOVA test has an \(\alpha\) chance of a Type I error (finding a difference between a pair that aren’t different),
- the overall Type I error rate will be much higher when running many tests simultaneously.
\[\begin{align} P(\text{making an error}) = & \alpha\\ P(\text{not making an error}) = & 1-\alpha\\ P(\text{not making an error in m tests}) = & (1-\alpha)^m\\ P(\text{making at least 1 error in m tests}) = & 1-(1-\alpha)^m \end{align}\]
The Bonferroni Correction (1/2)
A very conservative (but very popular) approach is to divide the \(\alpha\) level by how many tests \(m\) are being done:
\[\alpha_{Bonf} = \frac{\alpha}{m}\]
- This is equivalent to multiplying the
p-values by m:
\[p\textrm{-value} < \alpha_{Bonf} = \frac{\alpha}{m}\] is the same as \[m \cdot (p\textrm{-value}) < \alpha\] The Bonferroni correction is popular since it’s very easy to implement.
- The plot below shows the likelihood of making at least one Type I error depending on how may tests are done.
- Notice the likelihood decreases very quickly
- Unfortunately the likelihood of a Type II error is increasing as well
- It becomes “harder” and harder to reject \(H_0\) if doing many tests.
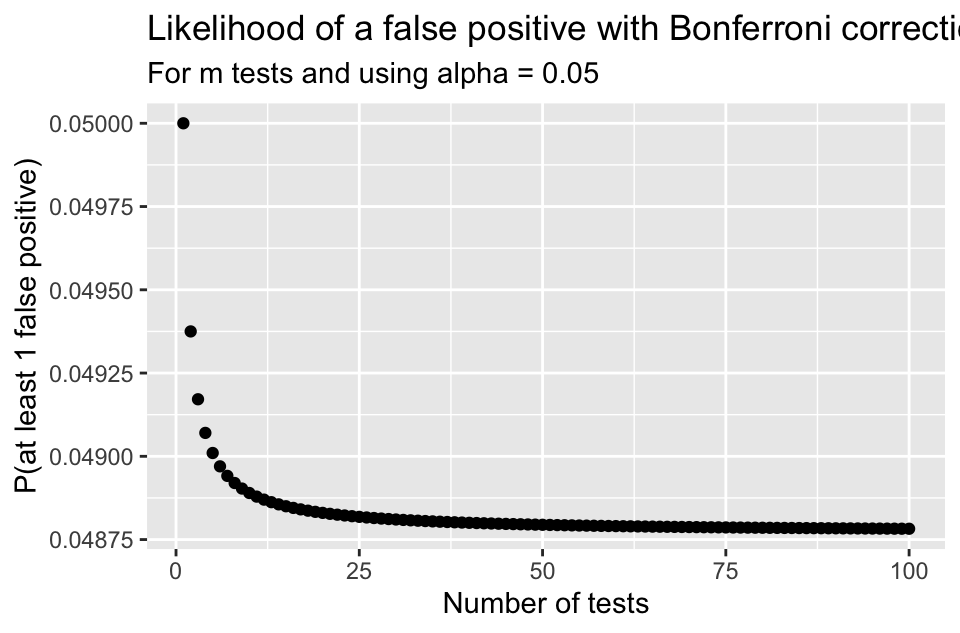
The Bonferroni Correction (2/2)
Pairwise t-tests without any p-value adjustments:
pairwise.t.test(employ$score,
employ$disability,
p.adj="none")
Pairwise comparisons using t tests with pooled SD
data: employ$score and employ$disability
none amputation crutches hearing
amputation 0.4477 - - -
crutches 0.1028 0.0184 - -
hearing 0.1732 0.5418 0.0035 -
wheelchair 0.4756 0.1433 0.3520 0.0401
P value adjustment method: none Pairwise t-tests with Bonferroni p-value adjustments:
pairwise.t.test(employ$score,
employ$disability,
p.adj="bonferroni")
Pairwise comparisons using t tests with pooled SD
data: employ$score and employ$disability
none amputation crutches hearing
amputation 1.000 - - -
crutches 1.000 0.184 - -
hearing 1.000 1.000 0.035 -
wheelchair 1.000 1.000 1.000 0.401
P value adjustment method: bonferroni - Since there were 10 tests, all the p-values were multiplied by 10.
- Are there any significant pairwise differences?
Tukey’s Honest Significance Test (HSD)
- Tukey’s Honest Significance Test (HSD) controls the “family-wise probability” of making a Type I error using a much less conservative method than Bonferroni
- It is specific to ANOVA
- In addition to adjusted p-values, it also calculates Tukey adjusted CI’s for all pairwise differences
- The function
TukeyHSD()creates a set of confidence intervals of the differences between means with the specified family-wise probability of coverage.
# need to run the model using `aov` instead of `lm`
empl_aov <- aov(score ~ disability, data = employ)
TukeyHSD(x=empl_aov, conf.level = 0.95) Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = score ~ disability, data = employ)
$disability
diff lwr upr p adj
amputation-none -0.4714286 -2.2031613 1.2603042 0.9399911
crutches-none 1.0214286 -0.7103042 2.7531613 0.4686233
hearing-none -0.8500000 -2.5817328 0.8817328 0.6442517
wheelchair-none 0.4428571 -1.2888756 2.1745899 0.9517374
crutches-amputation 1.4928571 -0.2388756 3.2245899 0.1232819
hearing-amputation -0.3785714 -2.1103042 1.3531613 0.9724743
wheelchair-amputation 0.9142857 -0.8174470 2.6460185 0.5781165
hearing-crutches -1.8714286 -3.6031613 -0.1396958 0.0277842
wheelchair-crutches -0.5785714 -2.3103042 1.1531613 0.8812293
wheelchair-hearing 1.2928571 -0.4388756 3.0245899 0.2348141plot(TukeyHSD(x=empl_aov, conf.level = 0.95))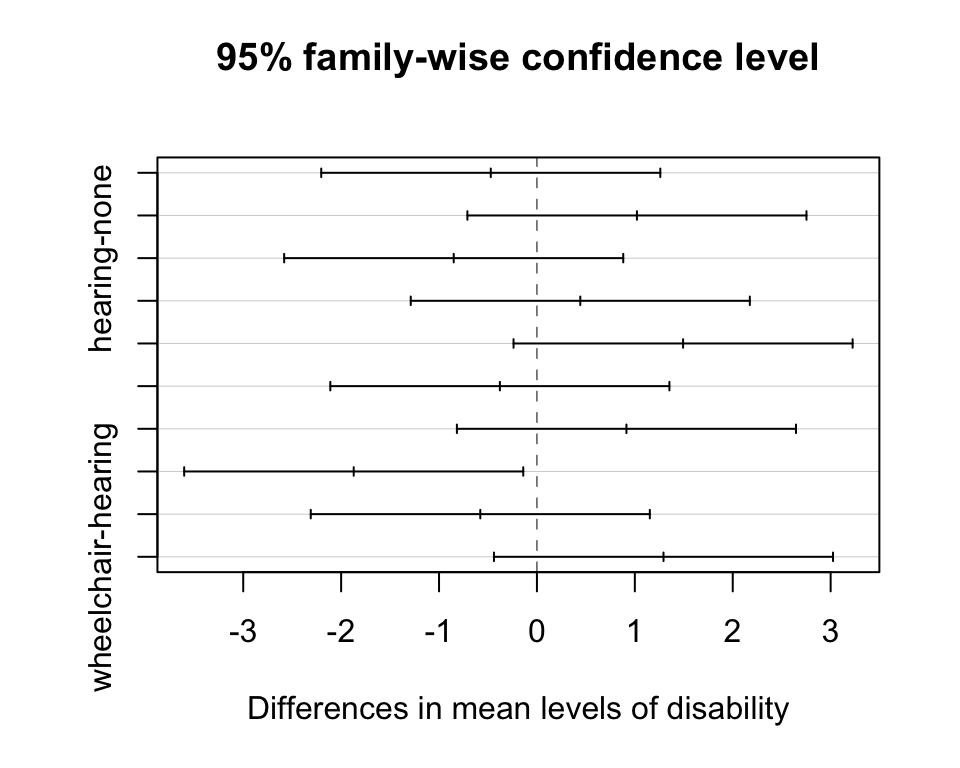
Visualization of pairwise CI’s
Which pair(s) of disabilities are significant after Tukey’s adjustments?
There are many more multiple testing adjustment procedures
- Bonferroni is popular because it’s so easy to apply
- Tukey’s HSD is usually used for ANOVA
- Code below used Holm’s adjustment
# default is Holm's adjustments
pairwise.t.test(employ$score,
employ$disability)
Pairwise comparisons using t tests with pooled SD
data: employ$score and employ$disability
none amputation crutches hearing
amputation 1.000 - - -
crutches 0.719 0.165 - -
hearing 0.866 1.000 0.035 -
wheelchair 1.000 0.860 1.000 0.321
P value adjustment method: holm - False discovery rate (fdr) p-value adjustments are popular in omics, or whenever there are many tests being run:
pairwise.t.test(employ$score,
employ$disability,
p.adj="fdr")
Pairwise comparisons using t tests with pooled SD
data: employ$score and employ$disability
none amputation crutches hearing
amputation 0.528 - - -
crutches 0.257 0.092 - -
hearing 0.289 0.542 0.035 -
wheelchair 0.528 0.287 0.503 0.134
P value adjustment method: fdr Multiple testing
post-hoc testing vs. testing many outcomes
Multiple testing: controlling the Type I error rate
- The multiple testing issue is not unique to ANOVA post-hoc testing.
- It is also a concern when running separate tests for many related outcomes.
- Beware of p-hacking!
Problem:
- Although one test has an \(\alpha\) chance of a Type I error (finding a difference between a pair that aren’t different),
- the overall Type I error rate will be much higher when running many tests simultaneously.
\[\begin{align} P(\text{making an error}) = & \alpha\\ P(\text{not making an error}) = & 1-\alpha\\ P(\text{not making an error in m tests}) = & (1-\alpha)^m\\ P(\text{making at least 1 error in m tests}) = & 1-(1-\alpha)^m \end{align}\]
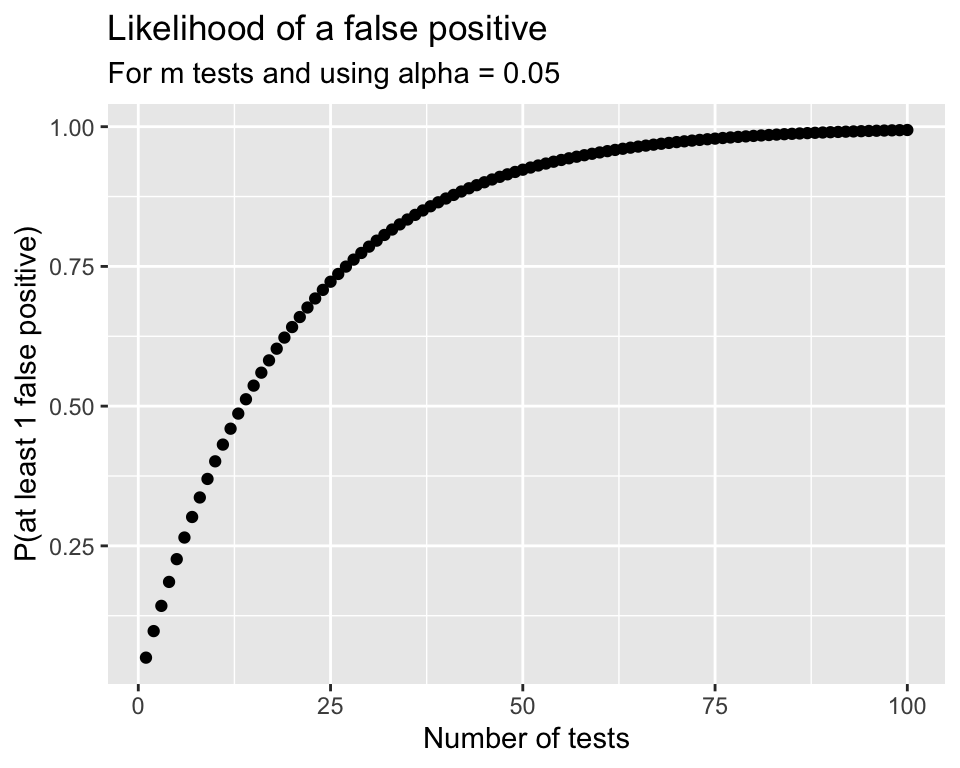
ANOVA Summary
\[\begin{align} H_0 &: \mu_1 = \mu_2 = ... = \mu_k\\ \text{vs. } H_A&: \text{At least one pair } \mu_i \neq \mu_j \text{ for } i \neq j \end{align}\]
- ANOVA table in R:
lm(score ~ disability, data = employ) %>% anova()Analysis of Variance Table
Response: score
Df Sum Sq Mean Sq F value Pr(>F)
disability 4 30.521 7.6304 2.8616 0.03013 *
Residuals 65 173.321 2.6665
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1ANOVA table
F-distribution & p-value

- Post-hoc testing