# run these every time you open Rstudio
library(tidyverse)
library(oibiostat)
library(janitor)
library(rstatix)
library(knitr)
library(gtsummary)
library(moderndive)
library(gt)
library(broom) # NEW!!
set.seed(456)Day 10 Part 2: Inference for mean difference from two-sample dependent/paired data (Section 5.2)
BSTA 511/611
Load packages
- Packages need to be loaded every time you restart R or render an Qmd file
- You can check whether a package has been loaded or not
- by looking at the Packages tab and
- seeing whether it has been checked off or not
Day 10 Part 2: Inference for mean difference from two-sample dependent/paired data (Section 5.2)
What we covered in Day 10 Part 1
(4.3, 5.1) Hypothesis testing for mean from one sample
- Introduce hypothesis testing using the case of analyzing a mean from one sample (group)
- Steps of a hypothesis test:
- level of significance
- null ( \(H_0\) ) and alternative ( \(H_A\) ) hypotheses
- test statistic
- p-value
- conclusion
- Run a hypothesis test in R
- Load a dataset - need to specify location of dataset
- R projects
- Run a t-test in R
tidy()the test output usingbroompackage
(4.3.3) Confidence intervals (CIs) vs. hypothesis tests
Goals for today: Part 2 - Class discussion
(5.2) Inference for mean difference from dependent/paired 2 samples
- Inference: CIs and hypothesis testing
- Exploratory data analysis (EDA) to visualize data
- Run paired t-test in R
One-sided CIs
Class discussion
- Inference for the mean difference from dependent/paired data is a special case of the inference for the mean from just one sample, that was already covered.
- Thus this part will be used for class discussion to practice CIs and hypothesis testing for one mean and apply it in this new setting.
- In class I will briefly introduce this topic, explain how it is similar and different from what we already covered, and let you work through the slides and code.
CI’s and hypothesis tests for different scenarios:
\[\text{point estimate} \pm z^*(or~t^*)\cdot SE,~~\text{test stat} = \frac{\text{point estimate}-\text{null value}}{SE}\]
| Day | Book | Population parameter |
Symbol | Point estimate | Symbol | SE |
|---|---|---|---|---|---|---|
| 10 | 5.1 | Pop mean | \(\mu\) | Sample mean | \(\bar{x}\) | \(\frac{s}{\sqrt{n}}\) |
| 10 | 5.2 | Pop mean of paired diff | \(\mu_d\) or \(\delta\) | Sample mean of paired diff | \(\bar{x}_{d}\) | ??? |
| 11 | 5.3 | Diff in pop means |
\(\mu_1-\mu_2\) | Diff in sample means |
\(\bar{x}_1 - \bar{x}_2\) | |
| 12 | 8.1 | Pop proportion | \(p\) | Sample prop | \(\widehat{p}\) | |
| 12 | 8.2 | Diff in pop proportions |
\(p_1-p_2\) | Diff in sample proportions |
\(\widehat{p}_1-\widehat{p}_2\) |
Steps in a Hypothesis Test
Set the level of significance \(\alpha\)
Specify the null ( \(H_0\) ) and alternative ( \(H_A\) ) hypotheses
- In symbols
- In words
- Alternative: one- or two-sided?
Calculate the test statistic.
Calculate the p-value based on the observed test statistic and its sampling distribution
Write a conclusion to the hypothesis test
- Do we reject or fail to reject \(H_0\)?
- Write a conclusion in the context of the problem
Examples of paired designs (two samples)
- Enroll pairs of identical twins to study a disease
- Enroll father & son pairs to study cholesterol levels
- Studying pairs of eyes
- Enroll people and collect data before & after an intervention (longitudinal data)
- Textbook example: Compare maximal speed of competitive swimmers wearing a wetsuit vs. wearing a regular swimsuit
- WIll use these data on homework
Come up with 2 more examples of paired study designs.
Can a vegetarian diet change cholesterol levels?
- Scenario:
- 24 non-vegetarian people were enrolled in a study
- They were instructed to adopt a vegetarian diet
- Cholesterol levels were measured before and after the diet
- Question: Is there evidence to support that cholesterol levels changed after the vegetarian diet?
- How to answer the question?
- First, calculate changes (differences) in cholesterol levels
- We usually do after - before if the data are longitudinal
- First, calculate changes (differences) in cholesterol levels
Calculate CI for the mean difference \(\delta\):
\[\bar{x}_d \pm t^*\cdot\frac{s_d}{\sqrt{n}}\]
Run a hypothesis test
Hypotheses
\[\begin{align} H_0:& \delta = \delta_0 \\ H_A:& \delta \neq \delta_0 \\ (or&~ <, >) \end{align}\]
Test statistic
\[ t_{\bar{x}_d} = \frac{\bar{x}_d - \delta_0}{\frac{s_d}{\sqrt{n}}} \]
EDA: Explore the cholesterol data
- Scenario:
- 24 non-vegetarian people were enrolled in a study
- They were instructed to adopt a vegetarian diet
- Cholesterol levels were measured before and after the diet
chol <- read_csv(here::here("data", "chol213.csv"))
glimpse(chol)Rows: 24
Columns: 2
$ Before <dbl> 195, 145, 205, 159, 244, 166, 250, 236, 192, 224, 238, 197, 169…
$ After <dbl> 146, 155, 178, 146, 208, 147, 202, 215, 184, 208, 206, 169, 182…chol %>%
get_summary_stats(type = "common") %>%
gt()| variable | n | min | max | median | iqr | mean | sd | se | ci |
|---|---|---|---|---|---|---|---|---|---|
| Before | 24 | 137 | 250 | 179 | 44.5 | 187.792 | 33.160 | 6.769 | 14.002 |
| After | 24 | 125 | 215 | 165 | 38.0 | 168.250 | 26.796 | 5.470 | 11.315 |
Make sure you are able to load the data on your computer!
EDA: Cholesterol levels before and after vegetarian diet
Describe the distributions of the before & after data.
ggplot(chol, aes(x=Before)) +
geom_density()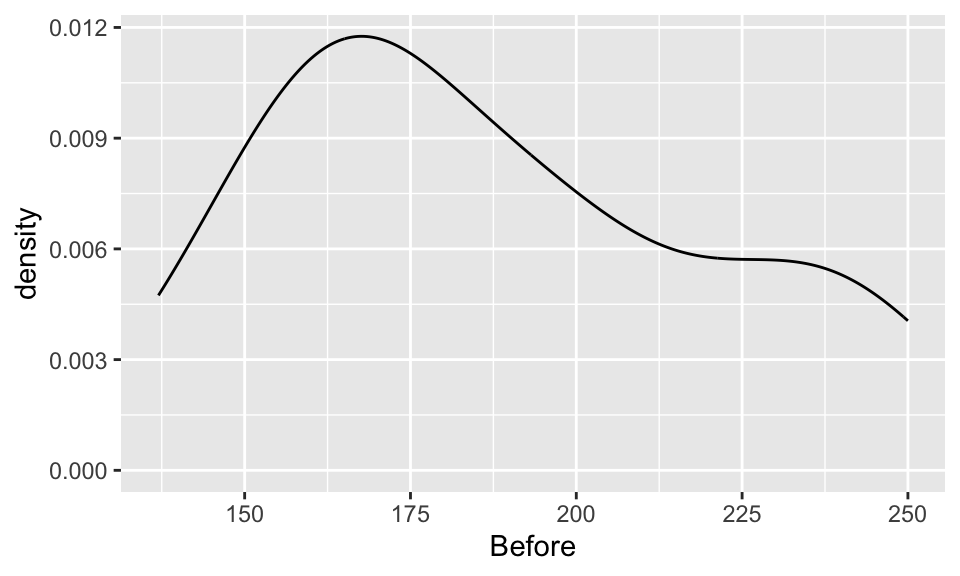
ggplot(chol, aes(x=Before)) +
geom_boxplot()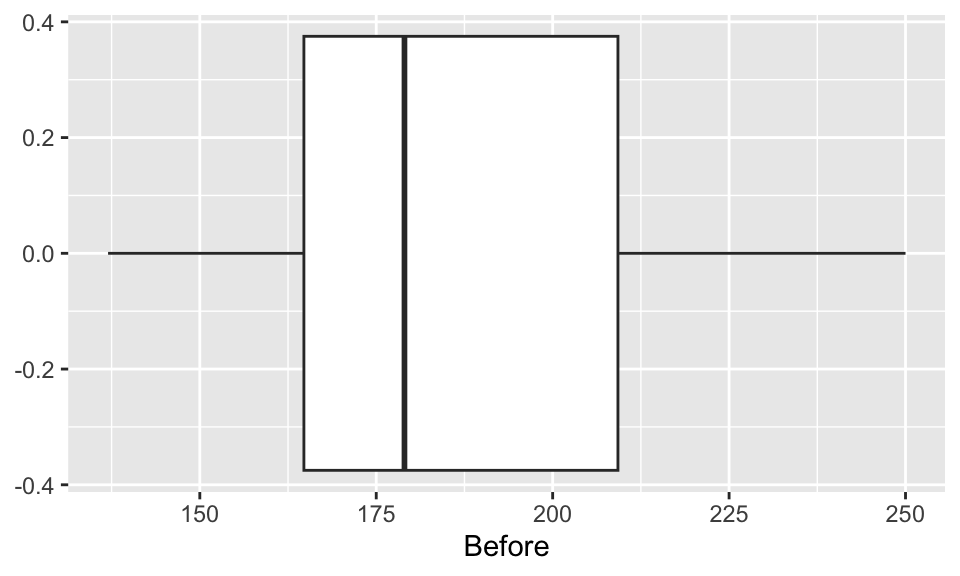
ggplot(chol, aes(x=After)) +
geom_density()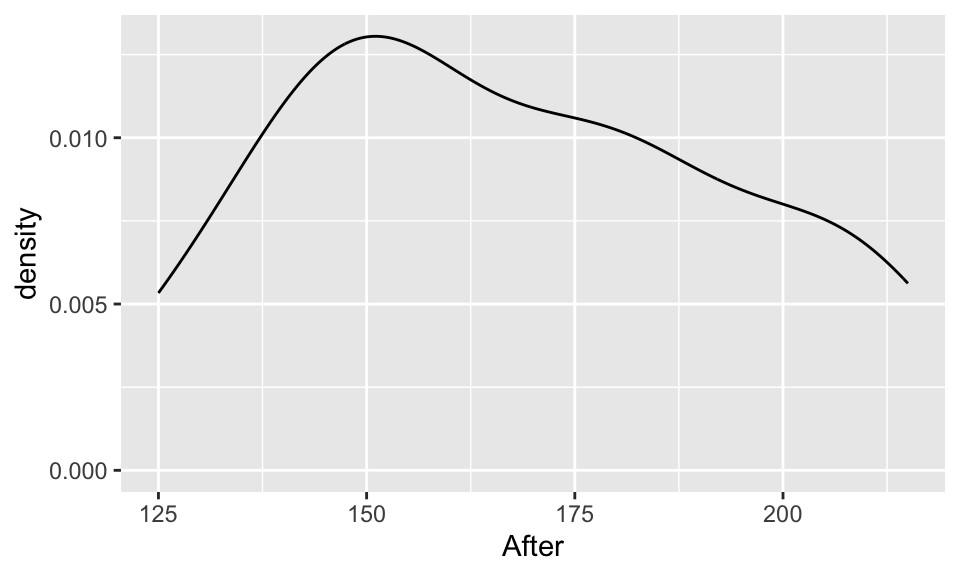
ggplot(chol, aes(x=After)) +
geom_boxplot()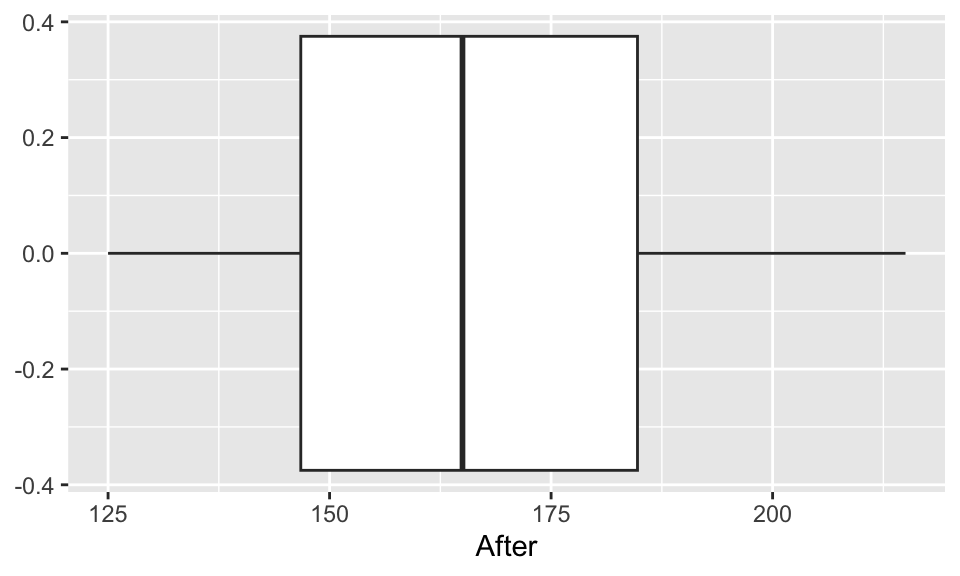
EDA: Spaghetti plot of cholesterol levels before & after diet
- Visualize the individual before vs. after diet changes in cholesterol levels
What does this figure tell us?
chol_long <- chol %>%
# need an ID column for the plot
# make it factor so that coloring is not on continuous scale
mutate(ID = factor(1:n())) %>%
# make data long for plot:
pivot_longer(
cols = Before:After,
names_to = "Time", # need a column for Before & After on x-axis
values_to = "Cholesterol") %>% # need a column of all cholesterol values for y-axis
mutate(
# change Time a factor variable so that can reorder
# levels so that Before is before After
Time = factor(Time, levels = c("Before", "After"))
)
ggplot(chol_long,
aes(x=Time, y = Cholesterol,
# need to include group = ID
# to create a line for each ID
color = ID, group = ID)) +
geom_line(show.legend = FALSE)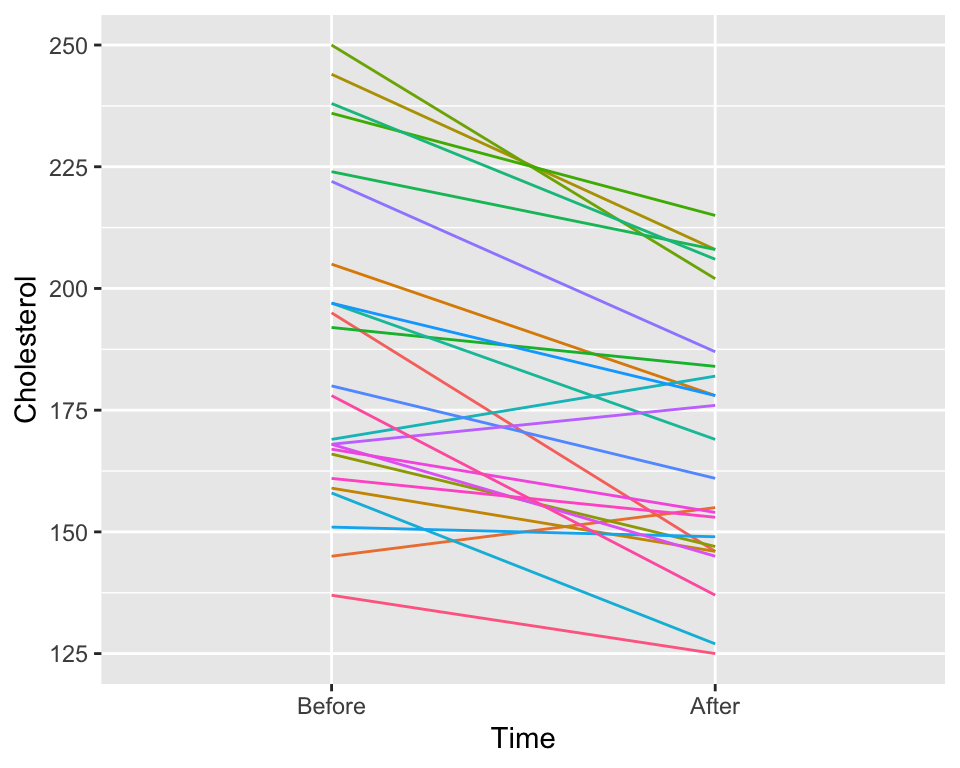
- You will not be expected to create this figure yourself.
EDA: Differences in cholesterol levels: After - Before diet
What is this code doing?
chol <- chol %>%
mutate(DiffChol = After-Before)
head(chol, 8)# A tibble: 8 × 3
Before After DiffChol
<dbl> <dbl> <dbl>
1 195 146 -49
2 145 155 10
3 205 178 -27
4 159 146 -13
5 244 208 -36
6 166 147 -19
7 250 202 -48
8 236 215 -21Is the mean of DiffChol the same as the difference in means of After - Before? Should it be? Why or why not?
chol %>%
get_summary_stats(type = "common") %>%
gt()| variable | n | min | max | median | iqr | mean | sd | se | ci |
|---|---|---|---|---|---|---|---|---|---|
| Before | 24 | 137 | 250 | 179 | 44.50 | 187.792 | 33.160 | 6.769 | 14.002 |
| After | 24 | 125 | 215 | 165 | 38.00 | 168.250 | 26.796 | 5.470 | 11.315 |
| DiffChol | 24 | -49 | 13 | -19 | 20.25 | -19.542 | 16.806 | 3.430 | 7.096 |
EDA: Differences in cholesterol levels: After - Before diet
Compare and contrast the 3 distributions. Comment on shape, center, and spread.
Before & After
ggplot(chol, aes(x=Before)) +
geom_density()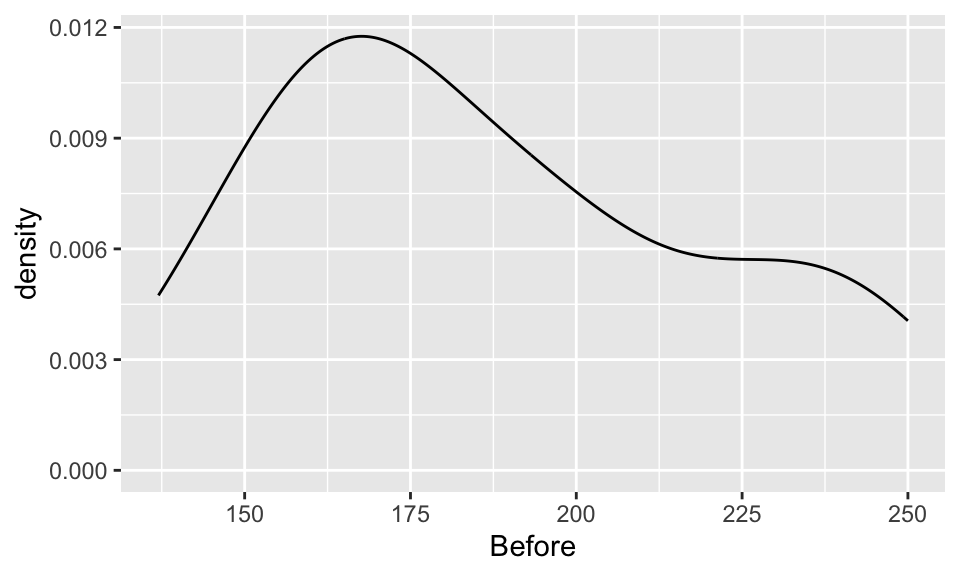
ggplot(chol, aes(x=After)) +
geom_density()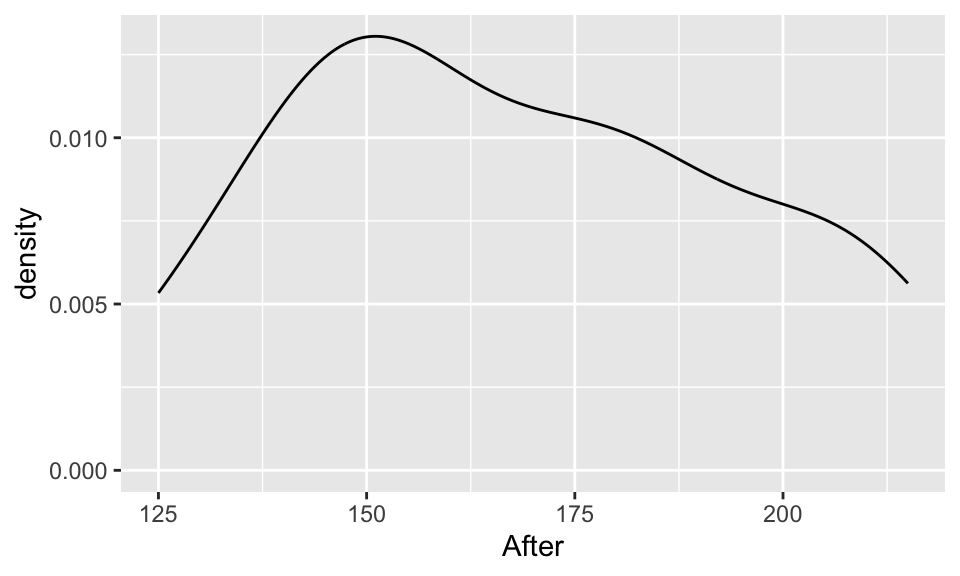
DiffChol
ggplot(chol, aes(x=DiffChol)) +
geom_density()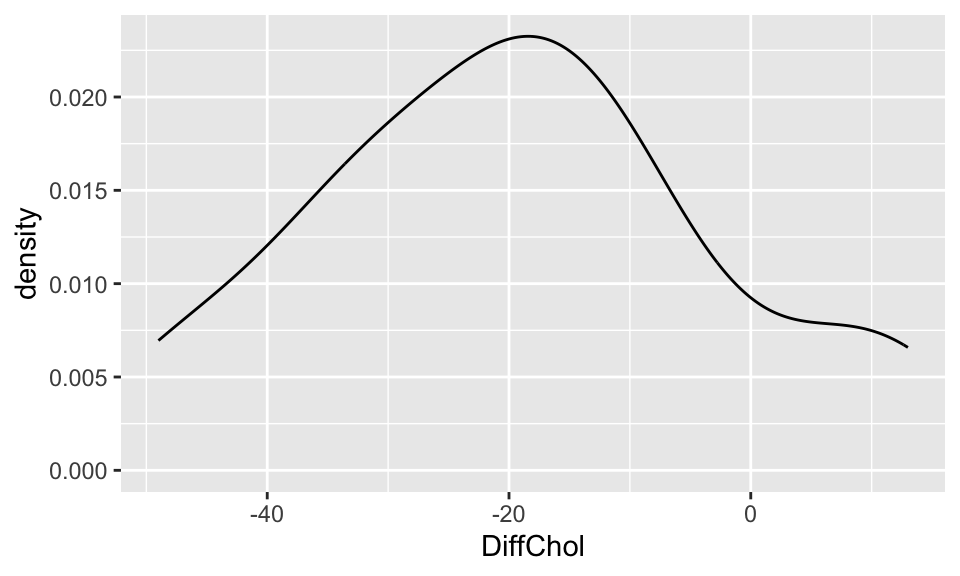
ggplot(chol, aes(x=DiffChol)) +
geom_boxplot()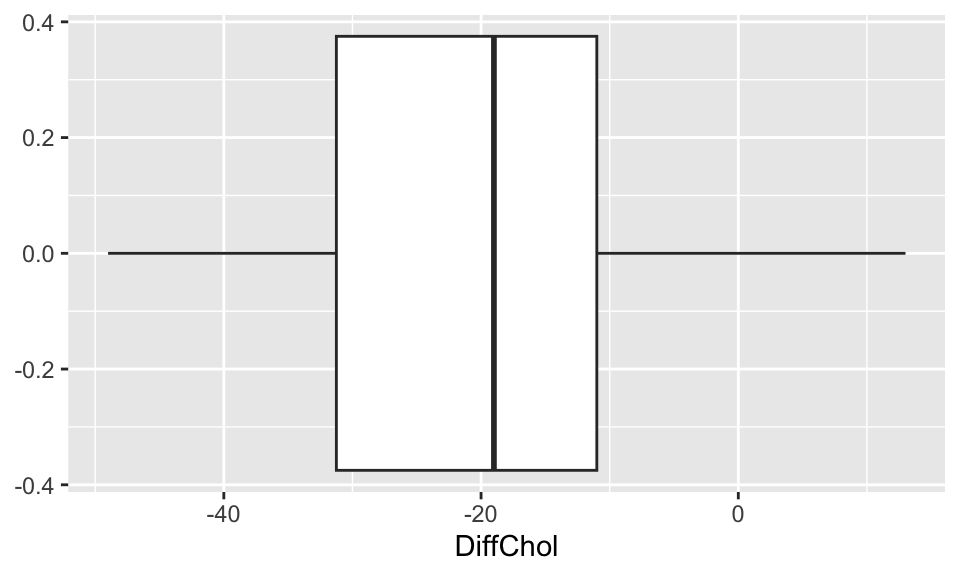
Steps in a Hypothesis Test
Set the level of significance \(\alpha\)
Specify the null ( \(H_0\) ) and alternative ( \(H_A\) ) hypotheses
- In symbols
- In words
- Alternative: one- or two-sided?
Calculate the test statistic.
Calculate the p-value based on the observed test statistic and its sampling distribution
Write a conclusion to the hypothesis test
- Do we reject or fail to reject \(H_0\)?
- Write a conclusion in the context of the problem
Step 2: Null & Alternative Hypotheses
- Question: Is there evidence to support that cholesterol levels changed after the vegetarian diet?
Null and alternative hypotheses in words Include as much context as possible
\(H_0\): The population mean difference in cholesterol levels after a vegetarian diet is fill in
\(H_A\): The population mean difference in cholesterol levels after a vegetarian diet is fill in
Null and alternative hypotheses in symbols
fill in the missing parts of the hypotheses.
\[~~~~H_0: \delta = \\ H_A: \delta \\\]
Step 3: Test statistic
chol %>% select(DiffChol) %>% get_summary_stats(type = "common") %>% gt()| variable | n | min | max | median | iqr | mean | sd | se | ci |
|---|---|---|---|---|---|---|---|---|---|
| DiffChol | 24 | -49 | 13 | -19 | 20.25 | -19.542 | 16.806 | 3.43 | 7.096 |
\[ t_{\bar{x}_d} = \frac{\bar{x}_d - \delta_0}{\frac{s_d}{\sqrt{n}}} \]
- Calculate the test statistic.
- Based on the value of the test statistic, do you think we are going to reject or fail to reject \(H_0\)?
- What probability distribution does the test statistic have?
- Are the assumptions for a paired t-test satisfied so that we can use the probability distribution to calculate the \(p\)-value??
n <- 24
alpha <- 0.05
mu <- 0
(p_area <- 1-alpha/2)[1] 0.975(xbar <- mean(chol$DiffChol))[1] -19.54167(sd <- sd(chol$DiffChol))[1] 16.80574(se <- sd/sqrt(n))[1] 3.430458(tstat <- (xbar - mu)/se)[1] -5.696519Step 4: p-value
The p-value is the probability of obtaining a test statistic just as extreme or more extreme than the observed test statistic assuming the null hypothesis \(H_0\) is true.
# specify upper and lower bounds of shaded region below
mu <- 0
std <- se
# The following figure is only an approximation of the
# sampling distribution since I used a normal instead
# of t-distribution to make it.
ggplot(data.frame(x = c(mu-6*std, mu+6*std)), aes(x = x)) +
stat_function(fun = dnorm,
args = list(mean = mu, sd = std)) +
scale_y_continuous(breaks = NULL) +
scale_x_continuous(breaks=c(mu, mu - 3.4*(1:6), mu + 3.4*(1:6))) +
theme(axis.text.x=element_text(angle = -30, hjust = 0)) +
labs(y = "",
x = "sample mean difference",
title = "Sampling distribution of mean difference") +
geom_vline(xintercept = c(-xbar, xbar),
color = "red")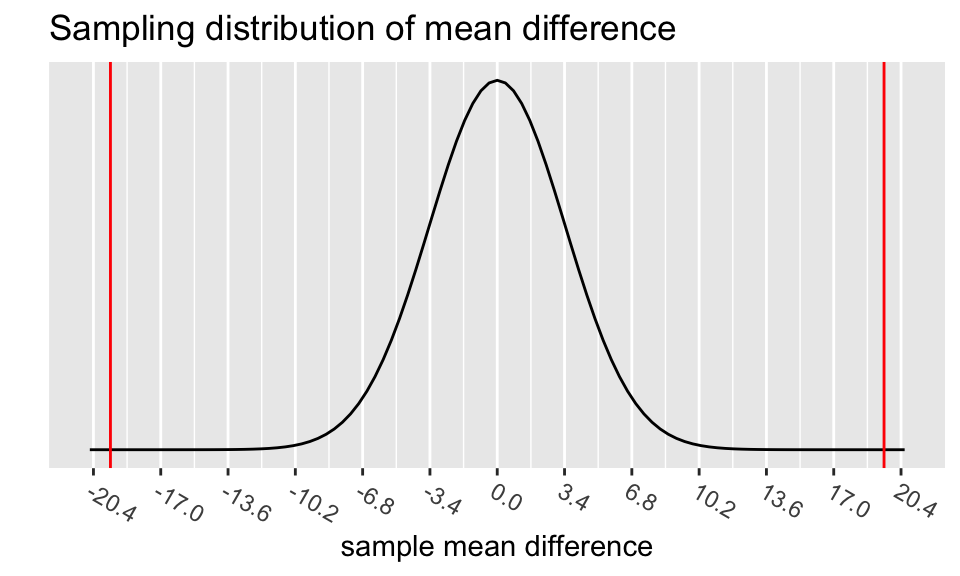
ggplot(data = data.frame(x = c(-6, 6)), aes(x)) +
stat_function(fun = dt, args = list(df = n-1)) +
ylab("") +
xlab("t-dist with df = 23") +
scale_y_continuous(breaks = NULL) +
scale_x_continuous(breaks=c(mu, mu - (1:5), mu + (1:5))) +
geom_vline(xintercept = c(-tstat,tstat),
color = "red")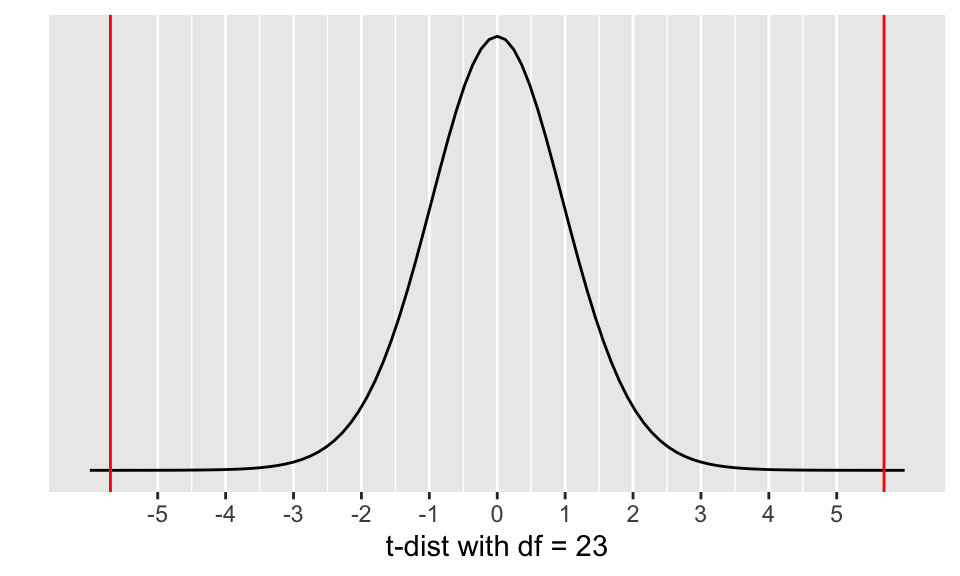
Calculate the p-value and shade in the area representing the p-value:
(pv <- 2*(pt(tstat, df=n-1)))[1] 8.434775e-06Step 5: Conclusion to hypothesis test
\[\begin{align} H_0:& \delta = 0 \\ H_A:& \delta \neq 0 \\ \end{align}\]
- Recall the \(p\)-value = \(8.434775 \cdot 10 ^{-6}\)
- Use \(\alpha\) = 0.05.
- Do we reject or fail to reject \(H_0\)?
Conclusion statement:
- Stats class conclusion
- There is sufficient evidence that the (population) mean difference in cholesterol levels after a vegetarian diet is different from 0 mg/dL ( \(p\)-value < 0.001).
- More realistic manuscript conclusion:
- After a vegetarian diet, cholesterol levels decreased by on average 19.54 mg/dL (SE = 3.43 mg/dL, 2-sided \(p\)-value < 0.001).
95% CI for the mean difference in cholesterol levels
chol %>% select(DiffChol) %>% get_summary_stats(type = "common") %>% gt()| variable | n | min | max | median | iqr | mean | sd | se | ci |
|---|---|---|---|---|---|---|---|---|---|
| DiffChol | 24 | -49 | 13 | -19 | 20.25 | -19.542 | 16.806 | 3.43 | 7.096 |
CI for \(\mu_d\) (or \(\delta\)):
n <- 24
alpha <- 0.05
(p_area <- 1-alpha/2)[1] 0.975(xbar <- mean(chol$DiffChol))[1] -19.54167(sd <- sd(chol$DiffChol))[1] 16.80574(tstar <- qt(p_area, df=n-1)) # df = n-1[1] 2.068658(se <- sd/sqrt(n))[1] 3.430458(moe <- tstar * se) [1] 7.096443(LB <- xbar - moe)[1] -26.63811(UB <- xbar + moe)[1] -12.44522\[\begin{align} \bar{x}_d &\pm t^*\cdot\frac{s_d}{\sqrt{n}}\\ -19.542 &\pm 2.069\cdot\frac{16.806}{\sqrt{24}}\\ -19.542 &\pm 2.069\cdot 3.43\\ -19.542 &\pm 7.096\\ (-26.638&, -12.445) \end{align}\]
Conclusion:
We are 95% that the (population) mean difference in cholesterol levels after a vegetarian diet is between -26.638 mg/dL and -12.445 mg/dL.
- Based on the CI, is there evidence the diet made a difference in cholesterol levels? Why or why not?
Running a paired t-test in R
R option 1: Run a 1-sample t.test using the paired differences
\(H_A: \delta \neq 0\)
t.test(x = chol$DiffChol, mu = 0)
One Sample t-test
data: chol$DiffChol
t = -5.6965, df = 23, p-value = 8.435e-06
alternative hypothesis: true mean is not equal to 0
95 percent confidence interval:
-26.63811 -12.44522
sample estimates:
mean of x
-19.54167 Run the code without mu = 0. Do the results change? Why or why not?
R option 2: Run a 2-sample t.test with paired = TRUE option
\(H_A: \delta \neq 0\)
- For a 2-sample t-test we specify both
x=andy= - Note:
mu = 0is the default value and doesn’t need to be specified
t.test(x = chol$Before, y = chol$After, mu = 0, paired = TRUE)
Paired t-test
data: chol$Before and chol$After
t = 5.6965, df = 23, p-value = 8.435e-06
alternative hypothesis: true mean difference is not equal to 0
95 percent confidence interval:
12.44522 26.63811
sample estimates:
mean difference
19.54167 What is different in the output compared to option 1?
R option 3: Run a 2-sample t.test with paired = TRUE option, but using the long data and a “formula”
- The data have to be in a
longformat for option 3, where each person has 2 rows: one for Before and one for After.- The long dataset
chol_longwas created for the slide “EDA: Spaghetti plot of cholesterol levels before & after diet”. - See the code to create it there.
- The long dataset
- What information is being stored in each of the columns?
# first 16 rows of long data:
head(chol_long, 16)# A tibble: 16 × 3
ID Time Cholesterol
<fct> <fct> <dbl>
1 1 Before 195
2 1 After 146
3 2 Before 145
4 2 After 155
5 3 Before 205
6 3 After 178
7 4 Before 159
8 4 After 146
9 5 Before 244
10 5 After 208
11 6 Before 166
12 6 After 147
13 7 Before 250
14 7 After 202
15 8 Before 236
16 8 After 215- Use the usual
t.test - What’s different is that
- instead of specifying the variables with
x=andy=, - we give a formula of the form
y ~ xusing just the variable names, - and then specify the name of the dataset using
data =
- instead of specifying the variables with
- This method is often used in practice, and more similar to the coding style of running a regression model (BSTA 512 & 513)
# using long data
# with columns Cholesterol & Time
t.test(Cholesterol ~ Time,
paired = TRUE,
data = chol_long)
Paired t-test
data: Cholesterol by Time
t = 5.6965, df = 23, p-value = 8.435e-06
alternative hypothesis: true mean difference is not equal to 0
95 percent confidence interval:
12.44522 26.63811
sample estimates:
mean difference
19.54167 - What is different in the output compared to option 1?
- Rerun the test using
Time ~ Cholesterol(switch the variables). What do you get?
Compare the 3 options
- How is the code similar and different for the 3 options?
- Given a dataset, how would you choose which of the 3 options to use?
# option 1
t.test(x = chol$DiffChol, mu = 0) %>% tidy() %>% gt() # tidy from broom package| estimate | statistic | p.value | parameter | conf.low | conf.high | method | alternative |
|---|---|---|---|---|---|---|---|
| -19.54167 | -5.696519 | 8.434775e-06 | 23 | -26.63811 | -12.44522 | One Sample t-test | two.sided |
# option 2
t.test(x = chol$Before, y = chol$After, mu = 0, paired = TRUE) %>% tidy() %>% gt()| estimate | statistic | p.value | parameter | conf.low | conf.high | method | alternative |
|---|---|---|---|---|---|---|---|
| 19.54167 | 5.696519 | 8.434775e-06 | 23 | 12.44522 | 26.63811 | Paired t-test | two.sided |
# option 3
t.test(Cholesterol ~ Time, paired = TRUE, data = chol_long) %>% tidy() %>% gt()| estimate | statistic | p.value | parameter | conf.low | conf.high | method | alternative |
|---|---|---|---|---|---|---|---|
| 19.54167 | 5.696519 | 8.434775e-06 | 23 | 12.44522 | 26.63811 | Paired t-test | two.sided |
What if we wanted to test whether the diet decreased cholesterol levels?
What changes in each of the steps?
Set the level of significance \(\alpha\)
Specify the hypotheses \(H_0\) and \(H_A\)
- Alternative: one- or two-sided?
Calculate the test statistic.
Calculate the p-value based on the observed test statistic and its sampling distribution
Write a conclusion to the hypothesis test
R: What if we wanted to test whether the diet decreased cholesterol levels?
- Which of the 3 options to run a paired t-test in R is being used below?
- How did the code change to account for testing a decrease in cholesterol levels?
- Which values in the output changed compared to testing for a change in cholesterol levels? How did they change?
# alternative = c("two.sided", "less", "greater")
t.test(x = chol$DiffChol, mu = 0, alternative = "less") %>% tidy() %>% gt()| estimate | statistic | p.value | parameter | conf.low | conf.high | method | alternative |
|---|---|---|---|---|---|---|---|
| -19.54167 | -5.696519 | 4.217387e-06 | 23 | -Inf | -13.6623 | One Sample t-test | less |
One-sided confidence intervals
Formula for a 2-sided (1- \(\alpha\) )% CI:
\[\bar{x} \pm t^*\cdot\frac{s}{\sqrt{n}}\] * \(t^*\) = qt(1-alpha/2, df = n-1) * \(\alpha\) is split over both tails of the distribution
A one-sided (1- \(\alpha\) )% CI has all (1- \(\alpha\) )% on just the left or the right tail of the distribution:
\[(\bar{x} - t^*\cdot\frac{s}{\sqrt{n}},~\infty) \\ (\infty,~\bar{x} + t^*\cdot\frac{s}{\sqrt{n}})\]
- \(t^*\) =
qt(1-alpha, df = n-1)for a 1-sided lower (1- \(\alpha\) )% CI - \(t^*\) =
qt(alpha, df = n-1)for a 1-sided upper (1- \(\alpha\) )% CI - A 1-sided CI gives estimates for a lower or upper bound of the population mean.
- See Section 4.2.3 of the V&H book for more
Today & what’s next?
CI’s and hypothesis tests for different scenarios:
\[\text{point estimate} \pm z^*(or~t^*)\cdot SE,~~\text{test stat} = \frac{\text{point estimate}-\text{null value}}{SE}\]
| Day | Book | Population parameter |
Symbol | Point estimate | Symbol | SE |
|---|---|---|---|---|---|---|
| 10 | 5.1 | Pop mean | \(\mu\) | Sample mean | \(\bar{x}\) | \(\frac{s}{\sqrt{n}}\) |
| 10 | 5.2 | Pop mean of paired diff | \(\mu_d\) or \(\delta\) | Sample mean of paired diff | \(\bar{x}_{d}\) | \(\frac{s_d}{\sqrt{n}}\) |
| 11 | 5.3 | Diff in pop means |
\(\mu_1-\mu_2\) | Diff in sample means |
\(\bar{x}_1 - \bar{x}_2\) | ??? |
| 12 | 8.1 | Pop proportion | \(p\) | Sample prop | \(\widehat{p}\) | |
| 12 | 8.2 | Diff in pop proportions |
\(p_1-p_2\) | Diff in sample proportions |
\(\widehat{p}_1-\widehat{p}_2\) |